Application: CyFinder
CyFinder – A Cytoscape Plugin to Find Subgraph Biomarkers in Biological Networks (Github, Tutorial).
In CyFinder, we implemented graph theoretic and community detection algorithms to discover subnetwork biomarkers from biological networks, such as gene co-expression and protein-protein interaction networks. The CyFinder plugin comes with functionality such as finding the cliques and bipartite graphs, which could be the building blocks for disease progression at the protein network level. CyFinder offers a Bipartite Layout (not available in the Cytoscape) in the Layout Menu that visualizes the nodes of a bipartite network in two disjoint sets. We implemented three renowned community detection algorithms (Edge Betweenness, Fastgreedy, and Walktrap), unavailable in other plugins. CyFinder can find the intersection of two or more graphs, and the resulting graphs could help identify the common network biomarkers in a cohort of patients for a particular disease. We also implemented two minimum spanning tree algorithms to identify both minimum and maximum spanning forests from a weighted graph, which could help analyze disease pathways.
The CyFinder App is a outcome of NSF CAREER project titled “NetDA—Protein Network-Based Software for Disease Analysis Using Cliques, Bipartite Graphs, and Diffusion Kernels.”
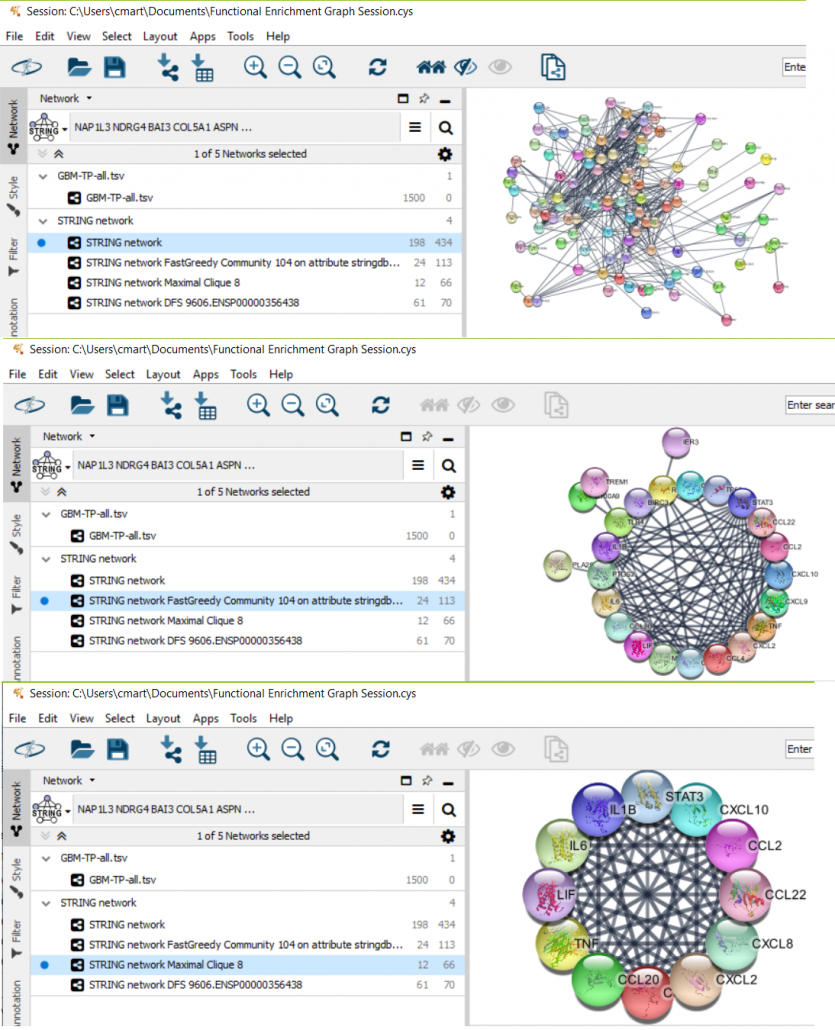
Sample CyFinder Results

